Taxonomic distribution of emerging species
Damiano Oldoni
Tim Adriaens
2024-07-03
This document follows the ranking pipeline to produce summary tables and graphs about the taxonomic distribution of the species defined as emerging (emerging status = 3).
1 Setup
Load libraries:
library(tidyverse) # To do data science
library(tidylog) # To provide feedback on dplyr functions
library(here) # To find files2 Get data
Read the tab separated files containing the emerging status output:
em_df <- readr::read_tsv(
here::here(
"data",
"output",
"ranking_emerging_status_hierarchical_strategy_Belgium.tsv"
),
na = ""
)Preview:
tidylog::slice_head(em_df, n = 10)3 Number of emerging species per class, year, location and variable
First, reshape the data:
emerging_df <-
em_df %>%
tidylog::pivot_longer(starts_with("year_"),
names_to = "variable",
values_to = "emerging_status"
)Preview:
tidylog::slice_head(emerging_df, n = 10)We calculate the number of emerging species
(emerging_status = 3) at class level for each combination
of:
- year
- variable: occupancy or number of occurrences
- area of interest: Belgium or protected areas
emerging_df <-
emerging_df %>%
tidylog::group_by(
.data$class,
.data$kingdom,
.data$variable
) %>%
tidylog::summarise(
n_emerging = length(.data$emerging_status[.data$emerging_status == 3]),
n_total = length(.data$emerging_status)
)Add year, variable and area of interest as columns:
emerging_df <-
emerging_df %>%
tidylog::mutate(
year = stringr::str_extract(.data$variable, pattern = "\\d+"),
area = stringr::str_extract(.data$variable, pattern = "Belgium$|natura2000$"),
variable = stringr::str_extract(.data$variable,
pattern = "occupancy|occs"
)
) %>%
tidylog::mutate(area = recode(.data$area, natura2000 = "Natura2000")) %>%
tidylog::relocate(n_emerging, n_total, .after = last_col()) %>%
tidylog::ungroup() %>%
dplyr::arrange(.data$year, desc(.data$n_emerging))Preview:
head(emerging_df, 20)Save this table as output file:
readr::write_tsv(
emerging_df,
here::here(
"data",
"output",
"number_of_emerging_taxa_per_class.tsv"
),
na = ""
)4 Summaries and graphs
Kingdoms with emerging species:
kingdoms <- emerging_df %>% tidylog::distinct(.data$kingdom)
kingdomsClasses with emerging species:
classes <- emerging_df %>% tidylog::distinct(.data$class, .data$kingdom)
classesDistribution of classes at kingdom levels:
classes %>%
tidylog::group_by(.data$kingdom) %>%
tidylog::count() %>%
dplyr::arrange(desc(.data$n))4.1 Taxonomic distribution at kingdom level
Number of emerging species at kingdom level:
emerging_df_kingdom <-
emerging_df %>%
tidylog::group_by(.data$kingdom, .data$variable, .data$year, .data$area) %>%
tidylog::summarise(
n_emerging = sum(.data$n_emerging),
n_total = sum(.data$n_total)
) %>%
tidylog::ungroup() %>%
tidylog::mutate(frac_emerging = .data$n_emerging / .data$n_total) %>%
dplyr::arrange(.data$variable, .data$area, .data$year, .data$kingdom)
emerging_df_kingdomShow the absolute distribution at kingdom level for each variable/area/year:
emerging_plot_kingdom <-
ggplot2::ggplot(emerging_df_kingdom) +
ggplot2::geom_col(aes(x = kingdom, y = n_emerging, fill = year),
position = "dodge"
) +
ggplot2::facet_grid(rows = vars(variable), cols = vars(area)) +
ggplot2::theme(
axis.text.x = ggplot2::element_text(
angle = 315,
hjust = 0,
vjust = 1
)
)
emerging_plot_kingdom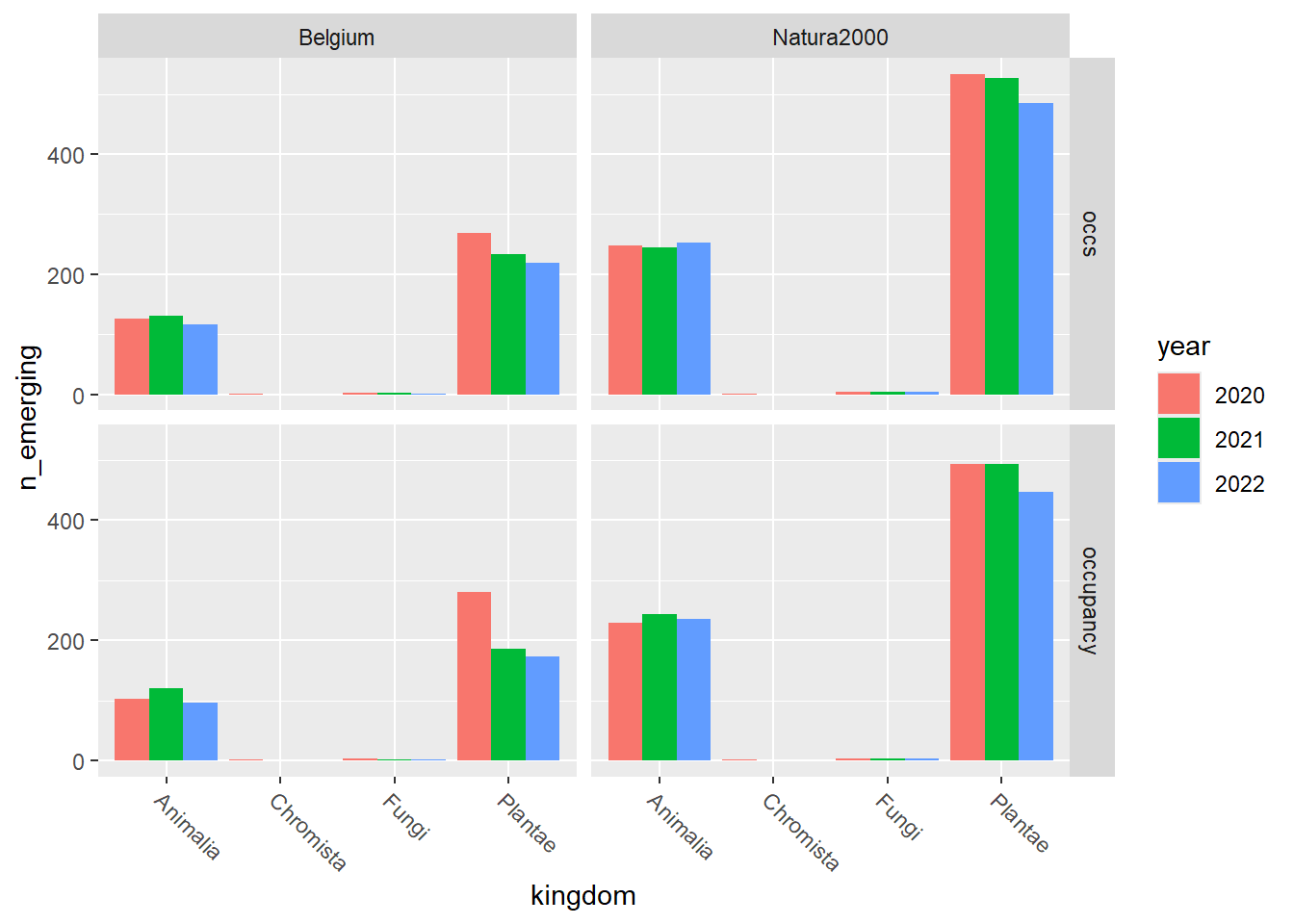
Show the relative distribution, i.e. the fraction number of emerging species / total number of alien species:
emerging_plot_kingdom_relative <-
ggplot2::ggplot(emerging_df_kingdom) +
ggplot2::geom_col(aes(x = kingdom, y = frac_emerging, fill = year),
position = "dodge"
) +
ggplot2::facet_grid(rows = vars(variable), cols = vars(area)) +
ggplot2::theme(
axis.text.x = ggplot2::element_text(
angle = 315,
hjust = 0,
vjust = 1
)
)
emerging_plot_kingdom_relative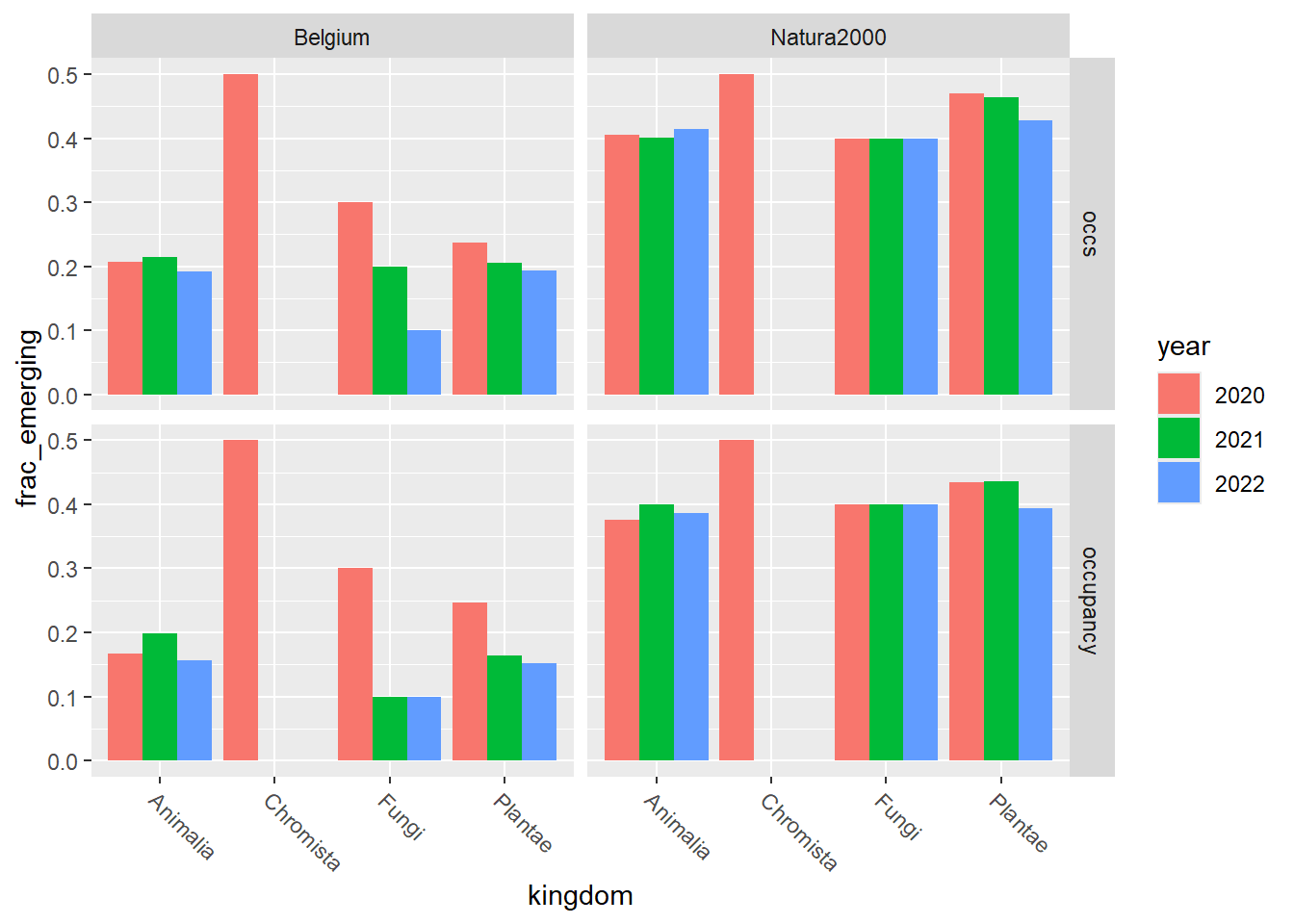
Save both the table emerging_df_kingdom and the graphs
with the absolute and relative distribution of emerging species at
kingdom level:
# save data.frame
readr::write_tsv(
x = emerging_df_kingdom,
file = here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"number_of_emerging_taxa_per_kingdom.tsv"
),
na = ""
)
# save plots with absolute distribution
ggplot2::ggsave(emerging_plot_kingdom,
filename = here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"n_emerging_taxa_per_kingdom.png"
)
)
# save plots with relative distribution
ggplot2::ggsave(emerging_plot_kingdom_relative,
filename = here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"n_emerging_taxa_per_kingdom_relative.png"
)
)4.2 Taxonomic distribution at class level
4.2.1 Occupancy: Belgium
Table:
emerging_df_BE_occupancy <-
emerging_df %>%
tidylog::filter(.data$area == "Belgium" & .data$variable == "occupancy")
emerging_df_BE_occupancyGraphs:
emerging_plots_BE_occupancy <- purrr::map(kingdoms$kingdom, function(x) {
data <-
emerging_df_BE_occupancy %>%
tidylog::filter(.data$kingdom == x & .data$n_emerging > 0)
ggplot2::ggplot(data) +
ggplot2::geom_col(aes(x = class, y = n_emerging, fill = year),
position = "dodge"
) +
ggplot2::theme(
axis.text.x = ggplot2::element_text(
angle = 315,
hjust = 0,
vjust = 1
)
)
})
emerging_plots_BE_occupancy## [[1]]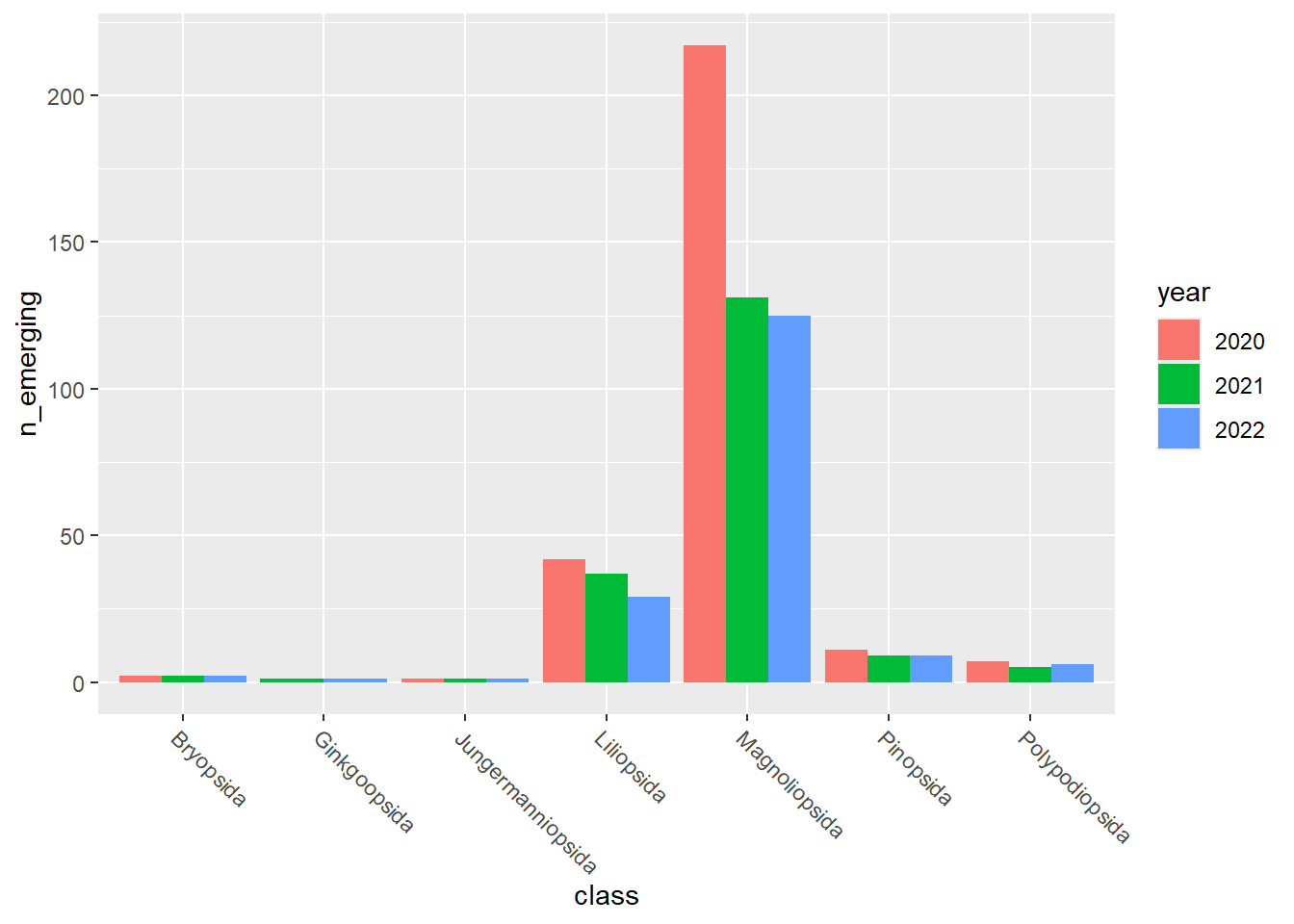
##
## [[2]]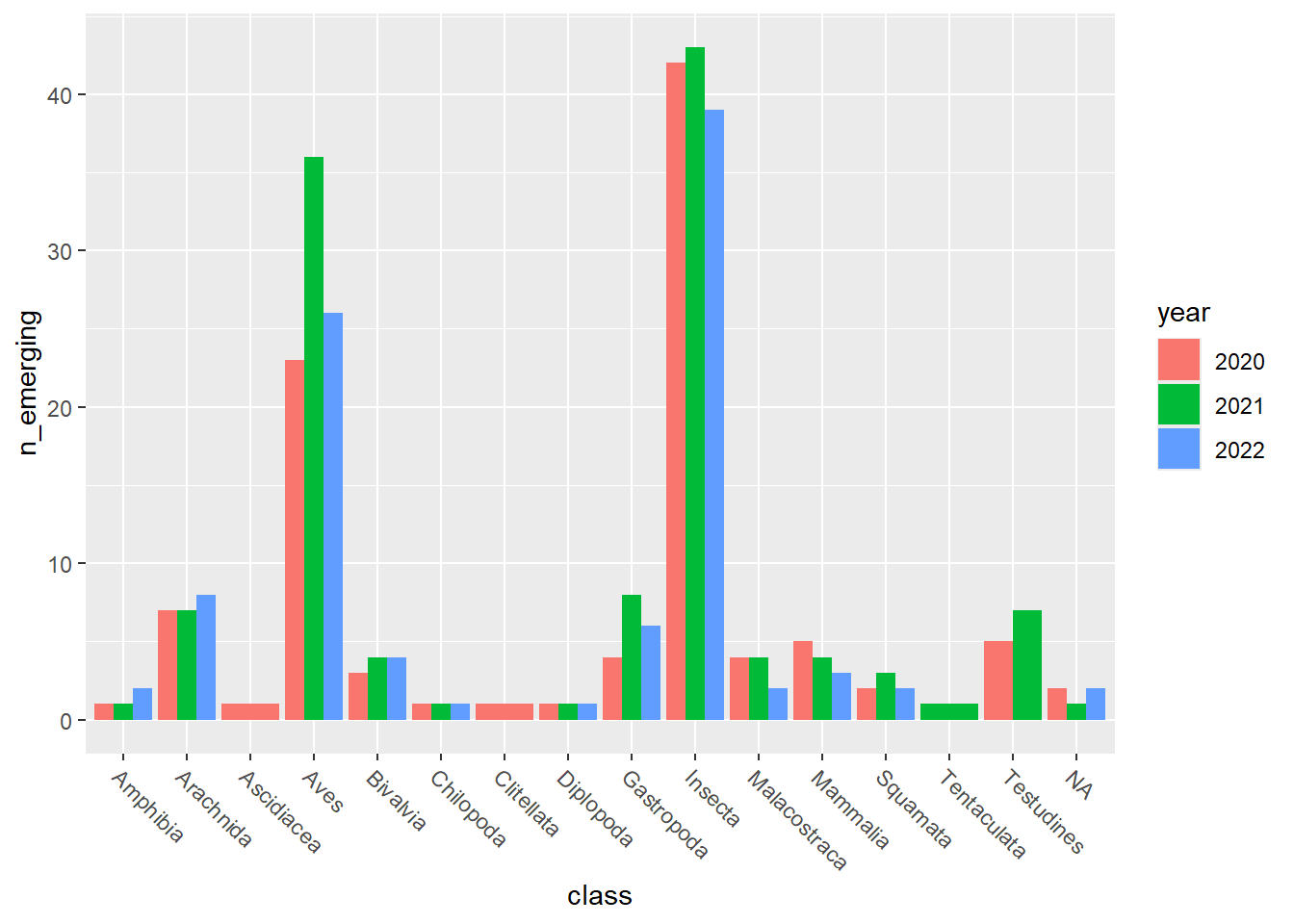
##
## [[3]]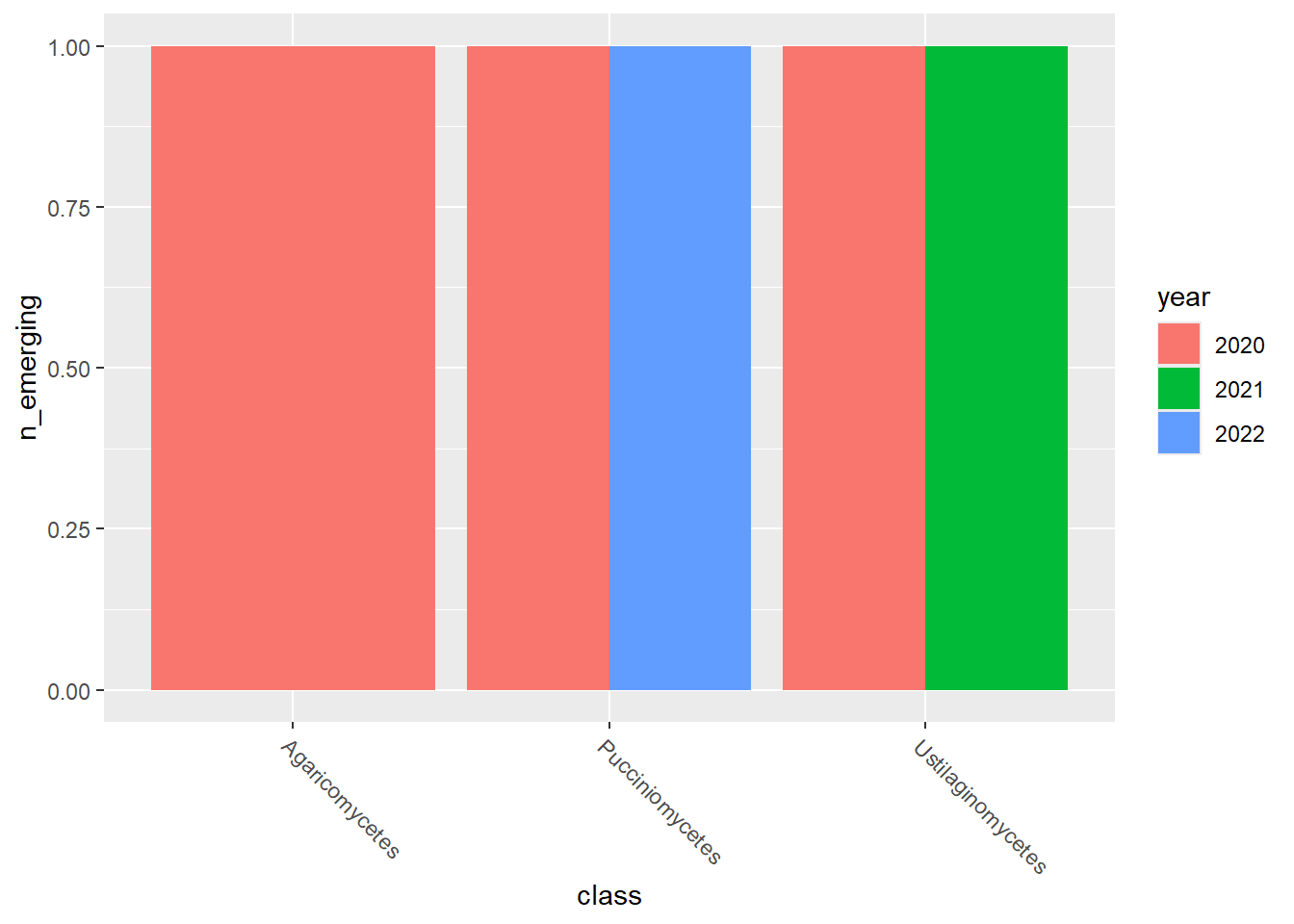
##
## [[4]]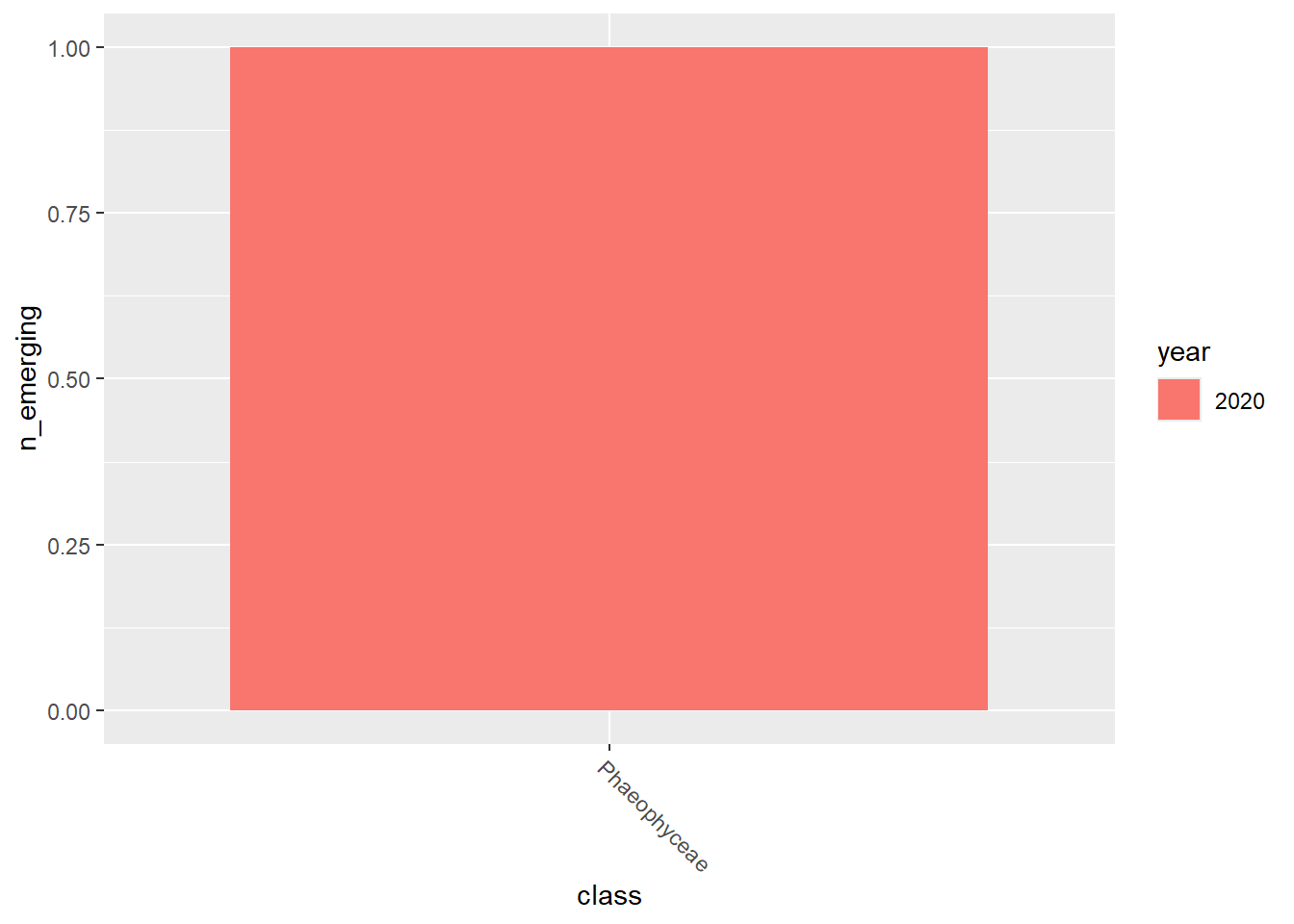
Save both table and graphs:
readr::write_tsv(
emerging_df_BE_occupancy,
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"n_emerging_taxa_per_class_in_BE_occupancy.tsv"
),
na = ""
)
purrr::walk2(
emerging_plots_BE_occupancy,
kingdoms$kingdom,
function(x, y) {
ggplot2::ggsave(
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
paste0(
"n_emerging_taxa_per_class_in_BE_occupancy_",
y,
".png"
)
),
x
)
}
)4.2.2 Occupancy: Natura2000
Table:
emerging_df_Natura2000_occupancy <-
emerging_df %>%
tidylog::filter(.data$area == "Natura2000" & .data$variable == "occupancy")
emerging_df_Natura2000_occupancyGraphs:
emerging_plots_Natura2000_occupancy <- purrr::map(
kingdoms$kingdom, function(x) {
data <- emerging_df_Natura2000_occupancy %>%
tidylog::filter(.data$kingdom == x & .data$n_emerging > 0)
ggplot2::ggplot(data) +
ggplot2::geom_col(aes(x = class, y = n_emerging, fill = year),
position = "dodge"
) +
ggplot2::theme(
axis.text.x = ggplot2::element_text(
angle = 315,
hjust = 0,
vjust = 1
)
)
}
)
emerging_plots_Natura2000_occupancy## [[1]]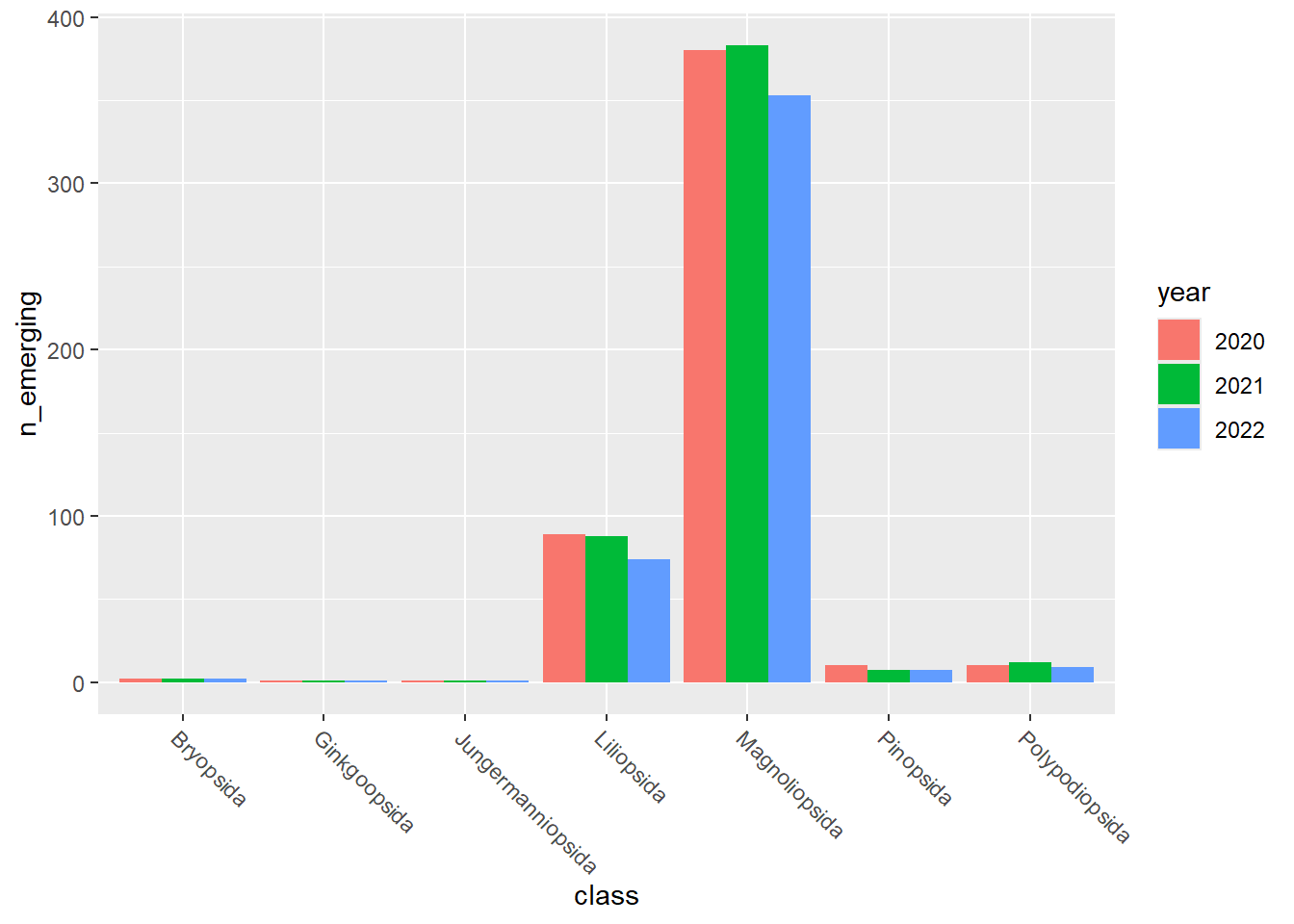
##
## [[2]]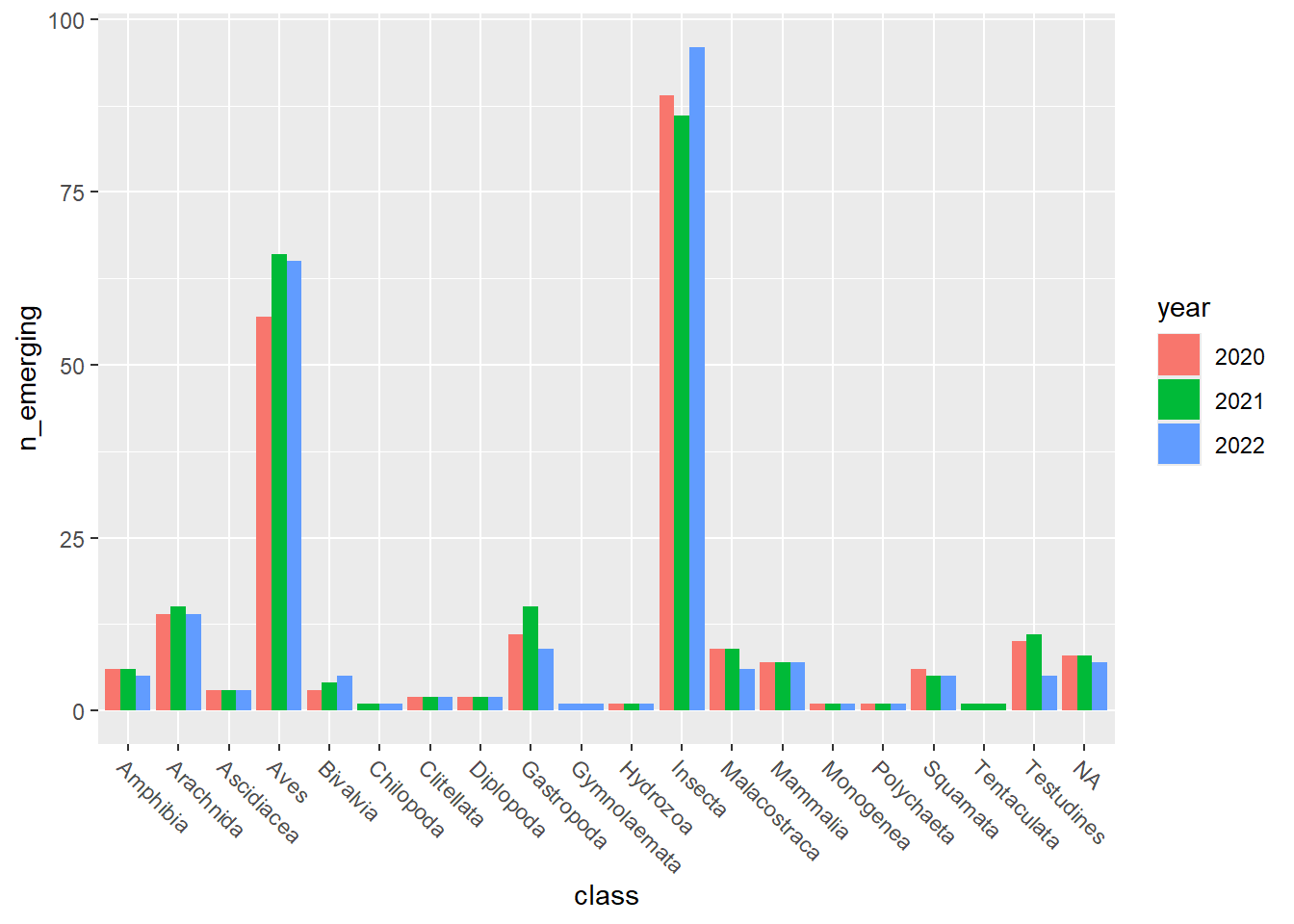
##
## [[3]]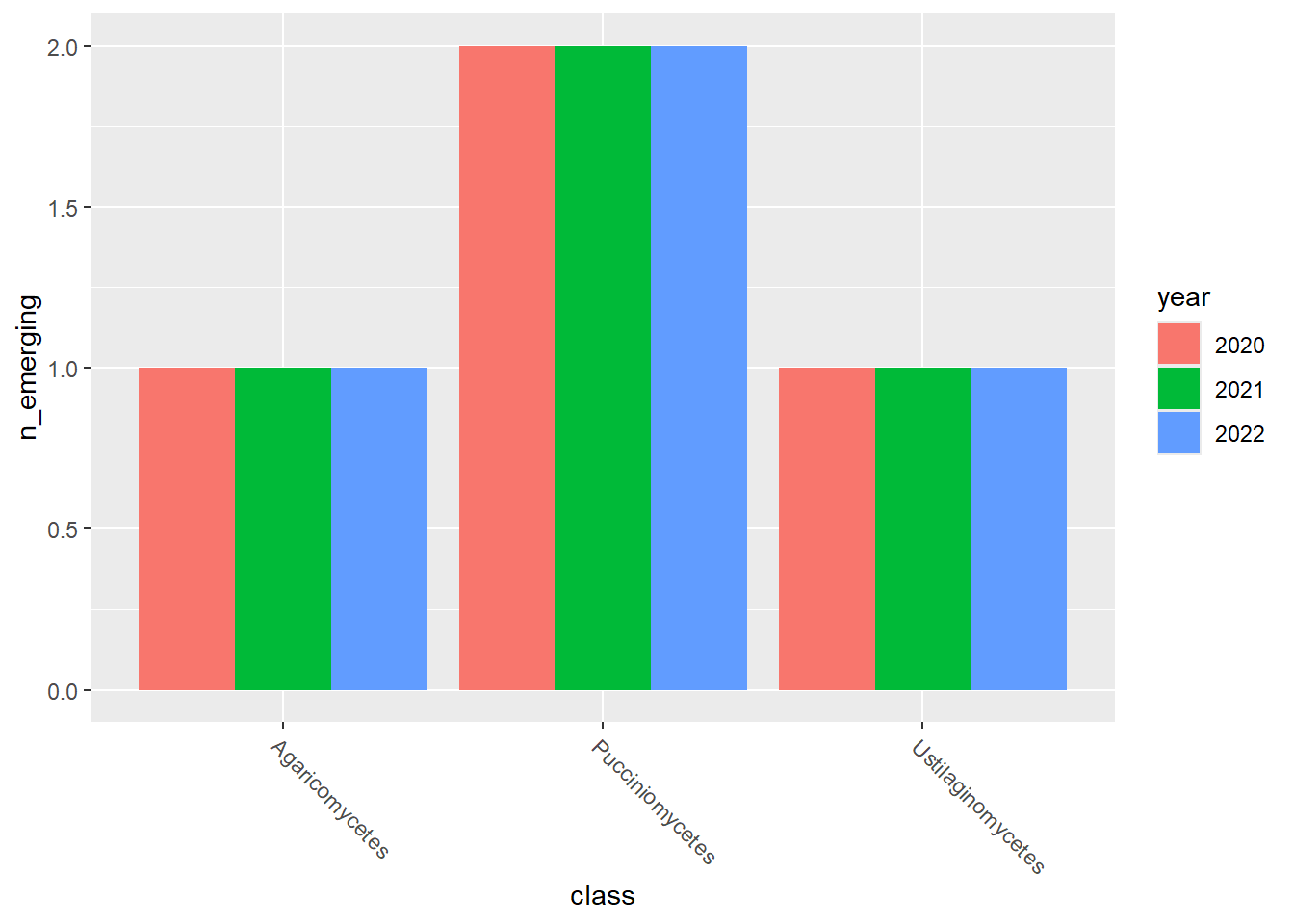
##
## [[4]]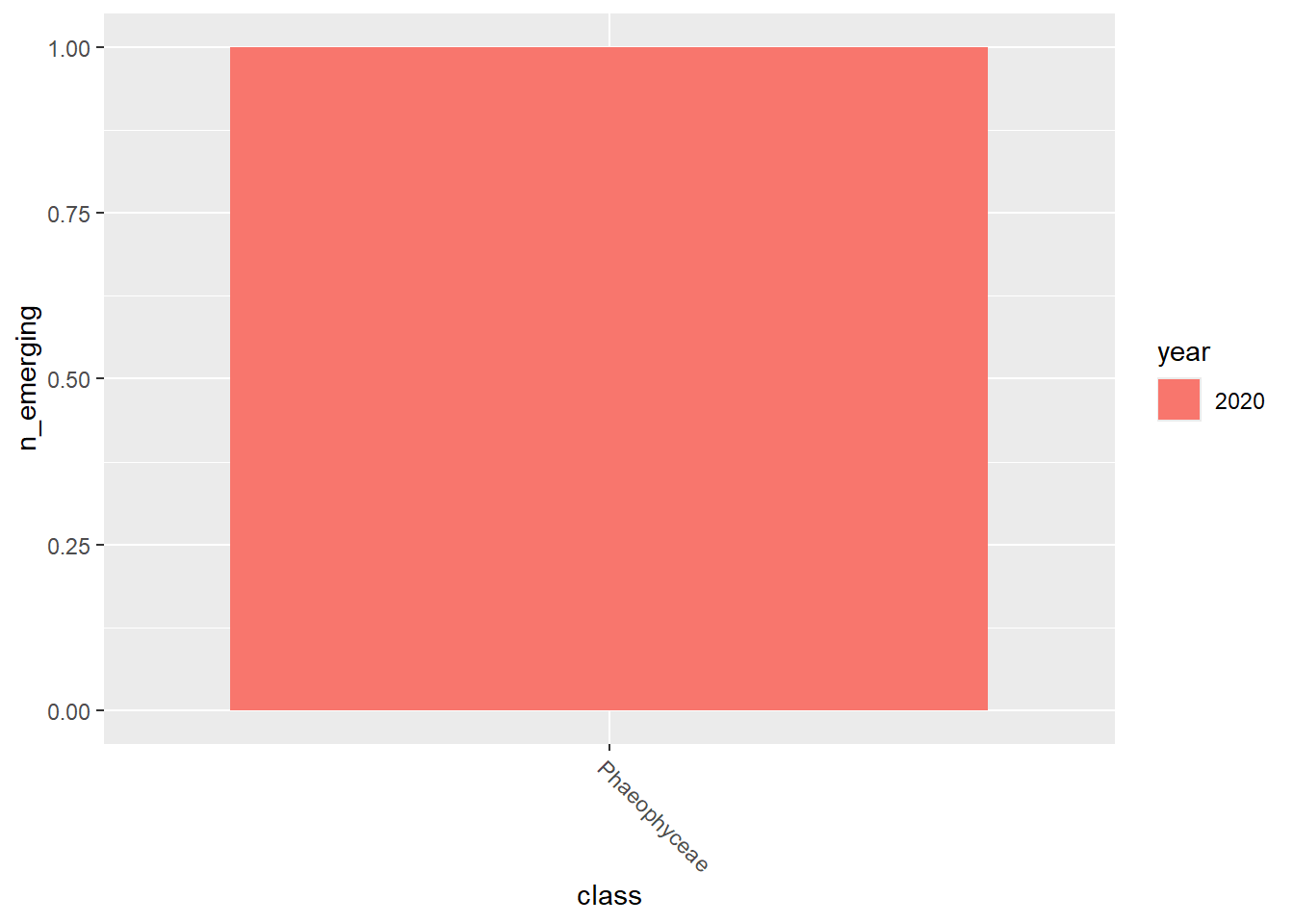
Save both table and graphs:
readr::write_tsv(
emerging_df_Natura2000_occupancy,
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"n_emerging_taxa_per_class_in_Natura2000_occupancy.tsv"
),
na = ""
)
purrr::walk2(
emerging_plots_Natura2000_occupancy,
kingdoms$kingdom,
function(x, y) {
ggplot2::ggsave(
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
paste0(
"n_emerging_taxa_per_class_in_Natura2000_occupancy_",
y, ".png"
)
),
x
)
}
)4.2.3 Number of occurrences: Belgium
Table:
emerging_df_BE_occs <-
emerging_df %>%
tidylog::filter(.data$area == "Belgium" & .data$variable == "occs")
emerging_df_BE_occsGraphs:
emerging_plots_BE_occs <- purrr::map(kingdoms$kingdom, function(x) {
data <- emerging_df_BE_occs %>%
tidylog::filter(.data$kingdom == x & .data$n_emerging > 0)
ggplot2::ggplot(data) +
ggplot2::geom_col(aes(x = class, y = n_emerging, fill = year),
position = "dodge"
) +
ggplot2::theme(
axis.text.x = ggplot2::element_text(
angle = 315,
hjust = 0,
vjust = 1
)
)
})
emerging_plots_BE_occs## [[1]]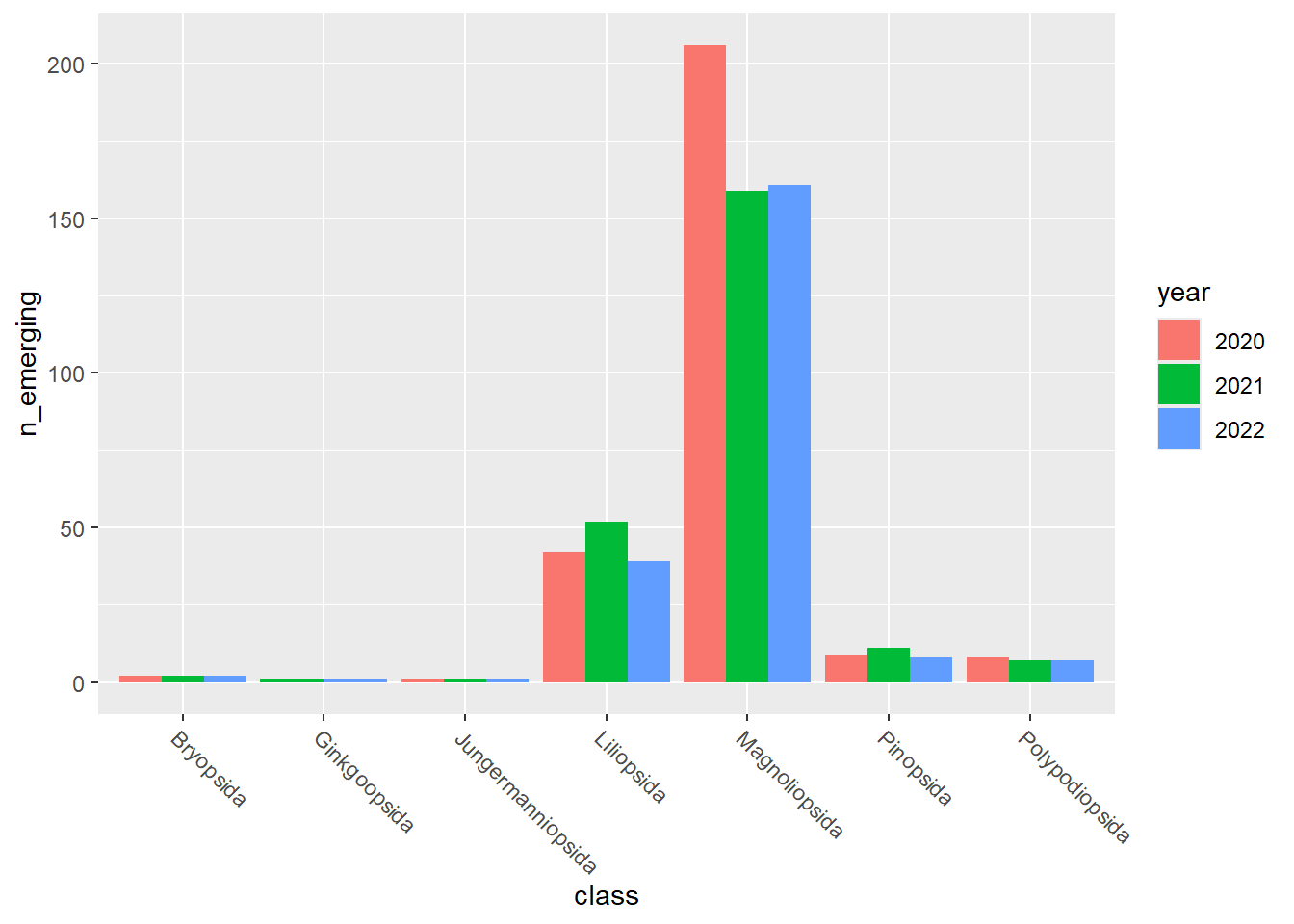
##
## [[2]]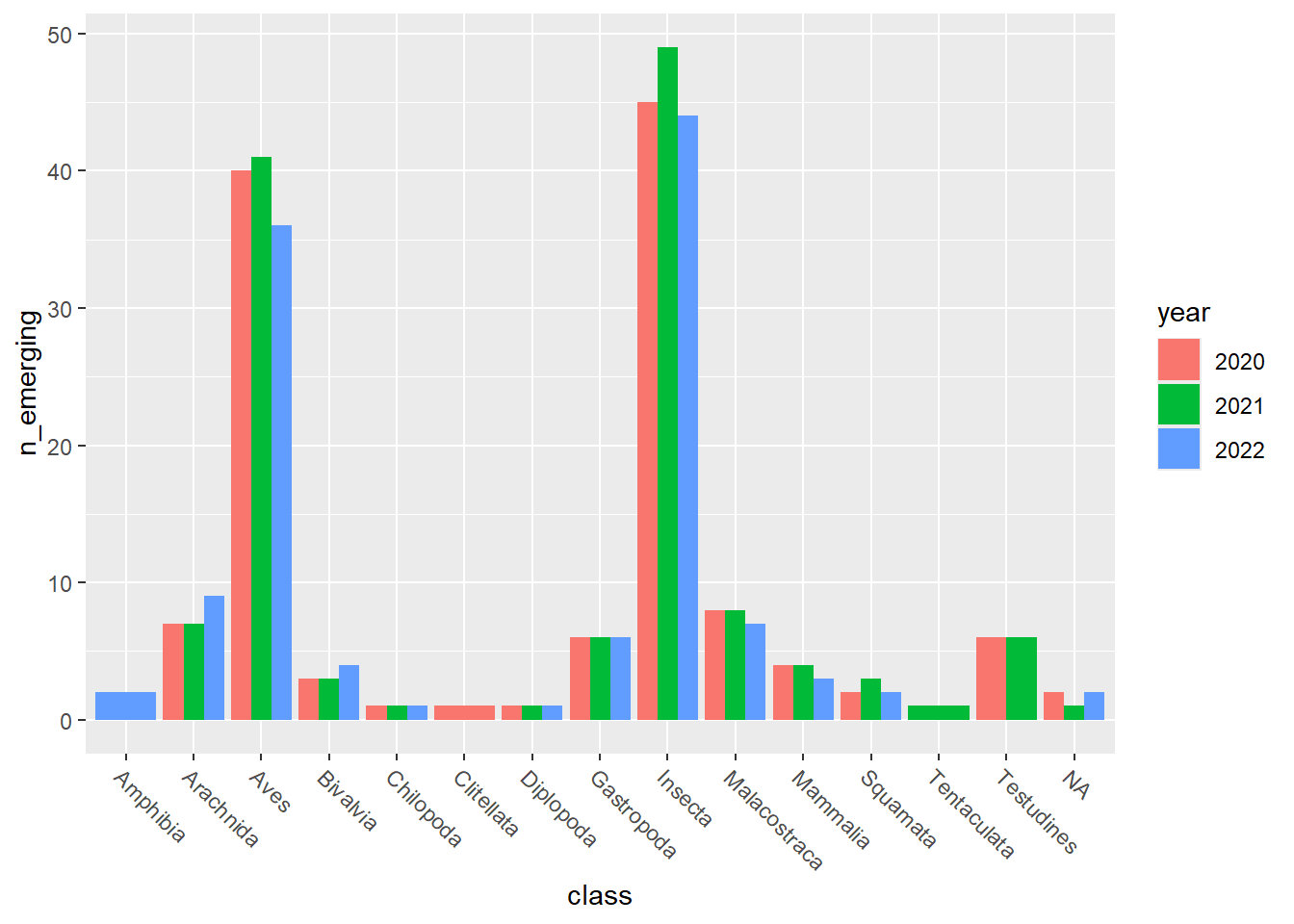
##
## [[3]]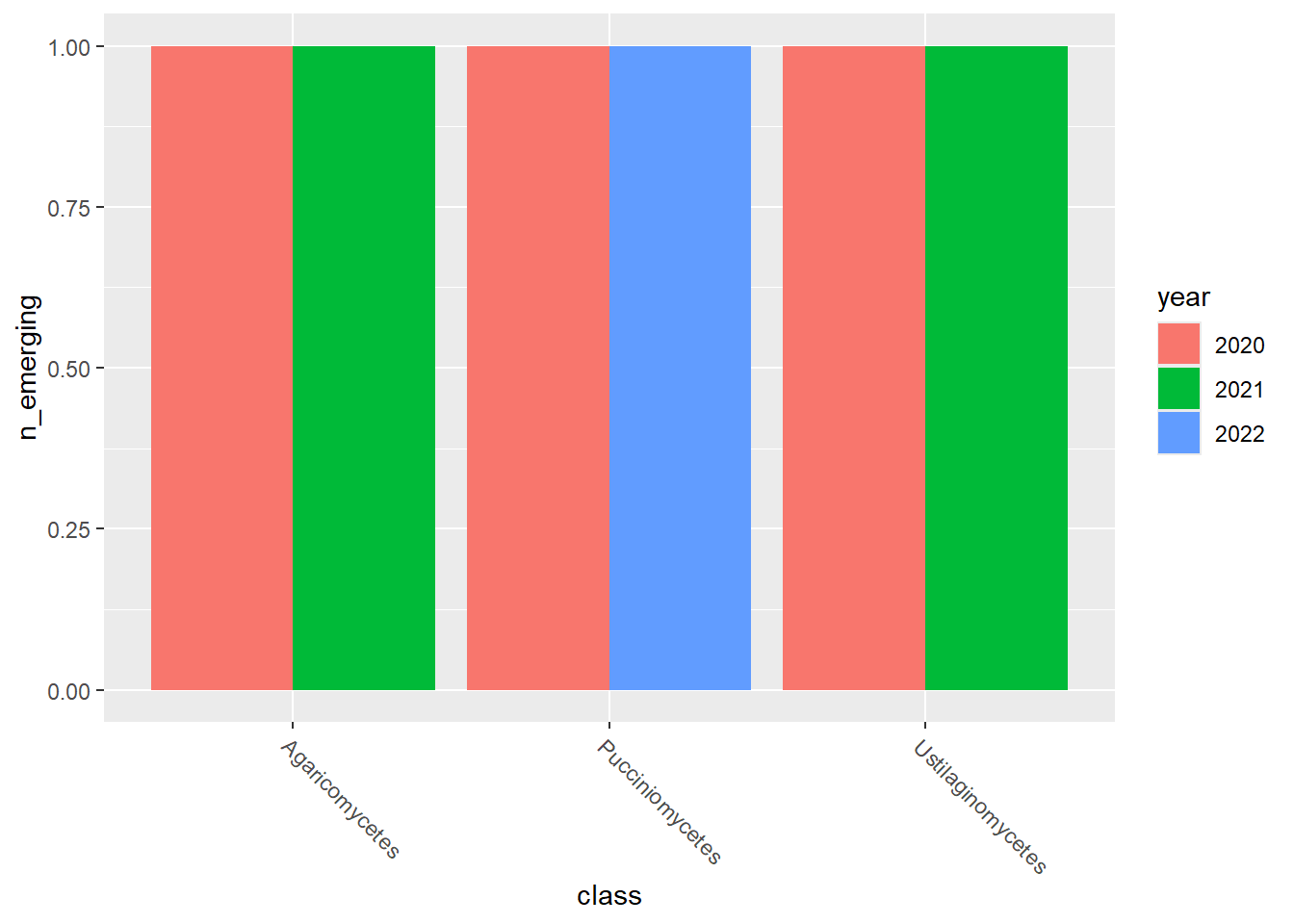
##
## [[4]]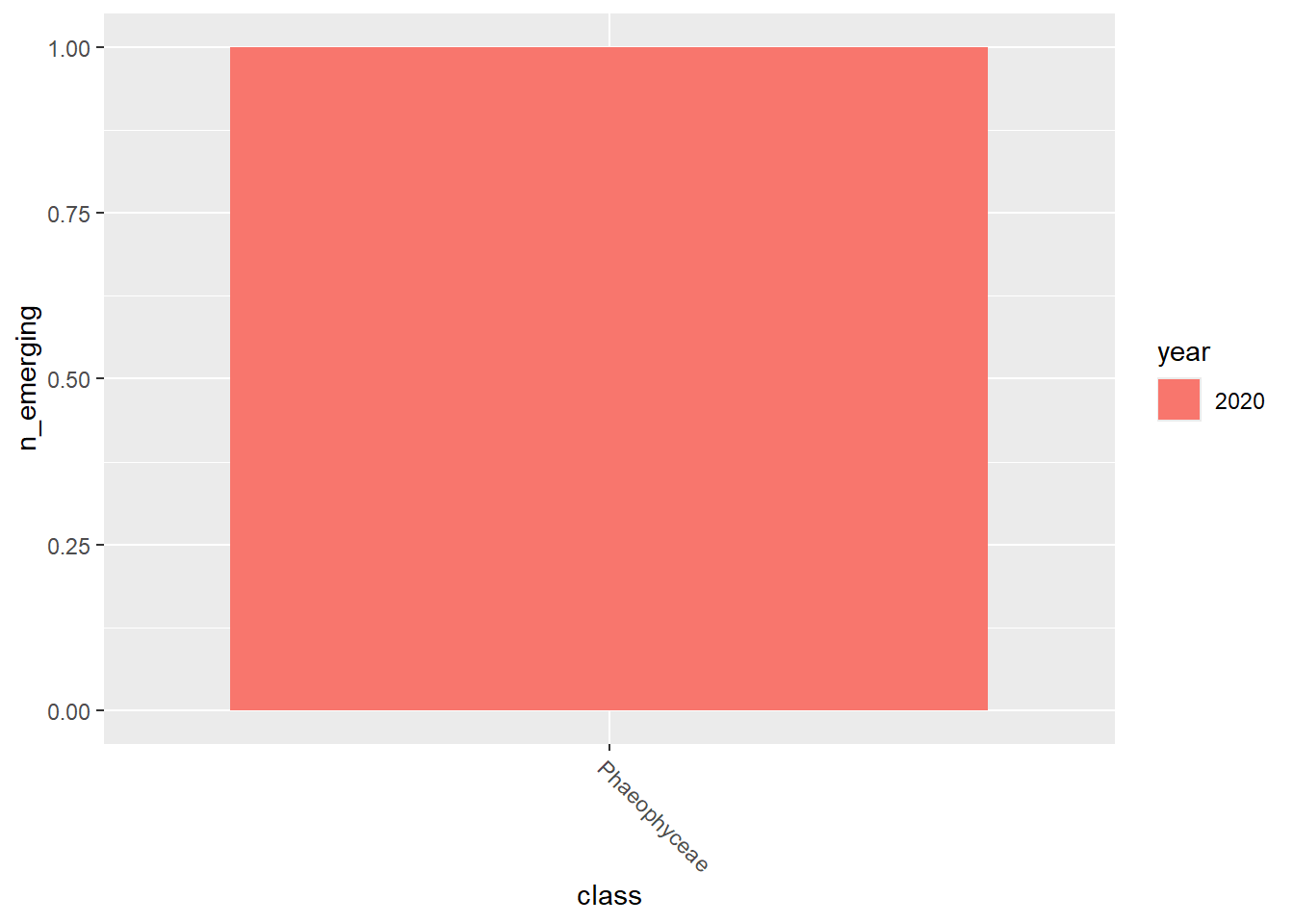
Save both table and graphs:
readr::write_tsv(
emerging_df_BE_occs,
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"n_emerging_taxa_per_class_in_Belgium_occs.tsv"
),
na = ""
)
purrr::walk2(
emerging_plots_BE_occs,
kingdoms$kingdom,
function(x, y) {
ggplot2::ggsave(
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
paste0(
"n_emerging_taxa_per_class_in_Belgium_occs_",
y, ".png"
)
),
x
)
}
)4.2.4 Number of occurrences: Natura2000
Table:
emerging_df_Natura2000_occs <-
emerging_df %>%
tidylog::filter(.data$area == "Natura2000" & .data$variable == "occs")
emerging_df_Natura2000_occsGraphs:
emerging_plots_Natura2000_occs <- purrr::map(kingdoms$kingdom, function(x) {
data <- emerging_df_Natura2000_occs %>%
tidylog::filter(.data$kingdom == x & .data$n_emerging > 0)
ggplot2::ggplot(data) +
ggplot2::geom_col(aes(x = class, y = n_emerging, fill = year),
position = "dodge"
) +
ggplot2::theme(
axis.text.x = ggplot2::element_text(
angle = 315,
hjust = 0,
vjust = 1
)
)
})
emerging_plots_Natura2000_occs## [[1]]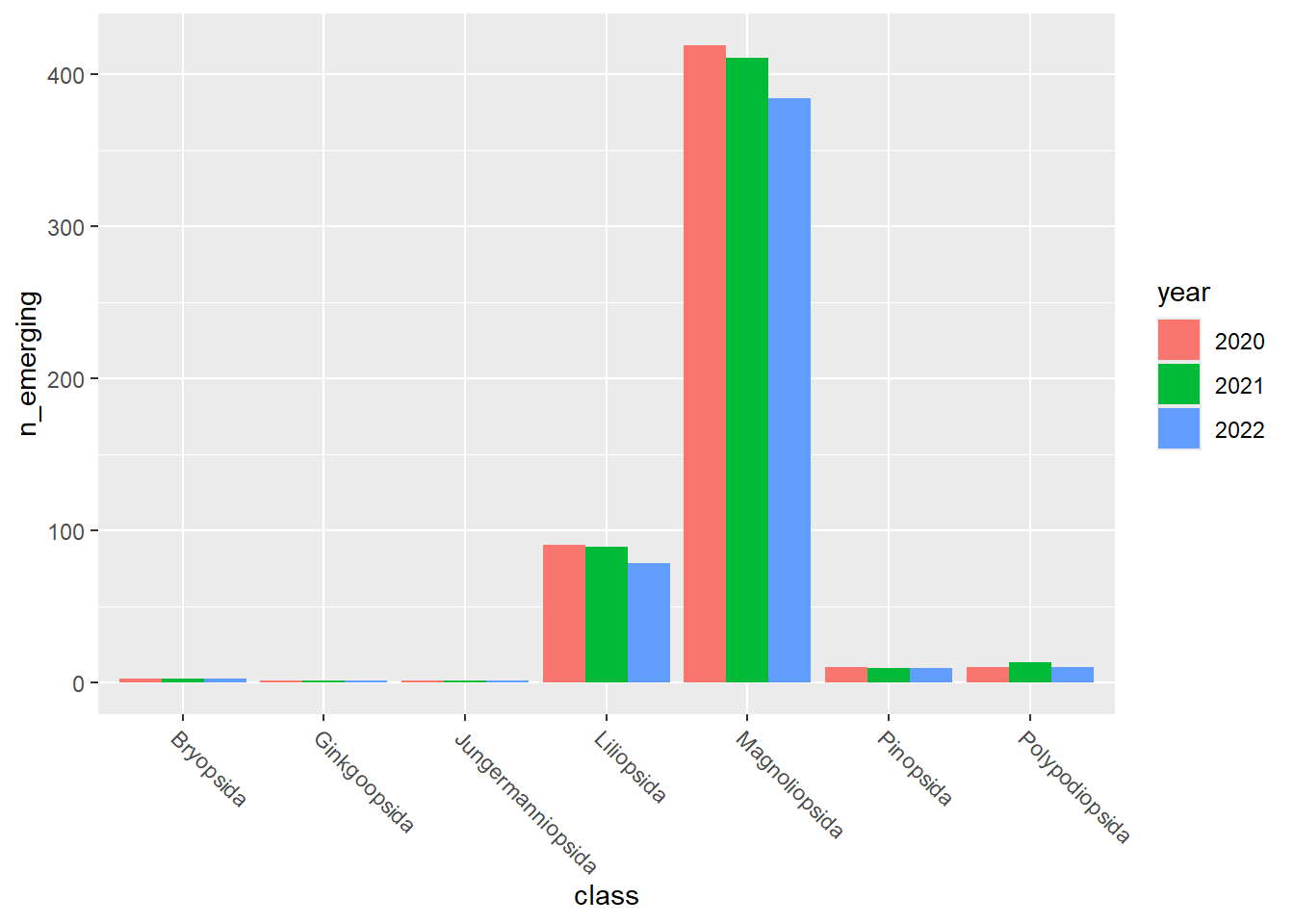
##
## [[2]]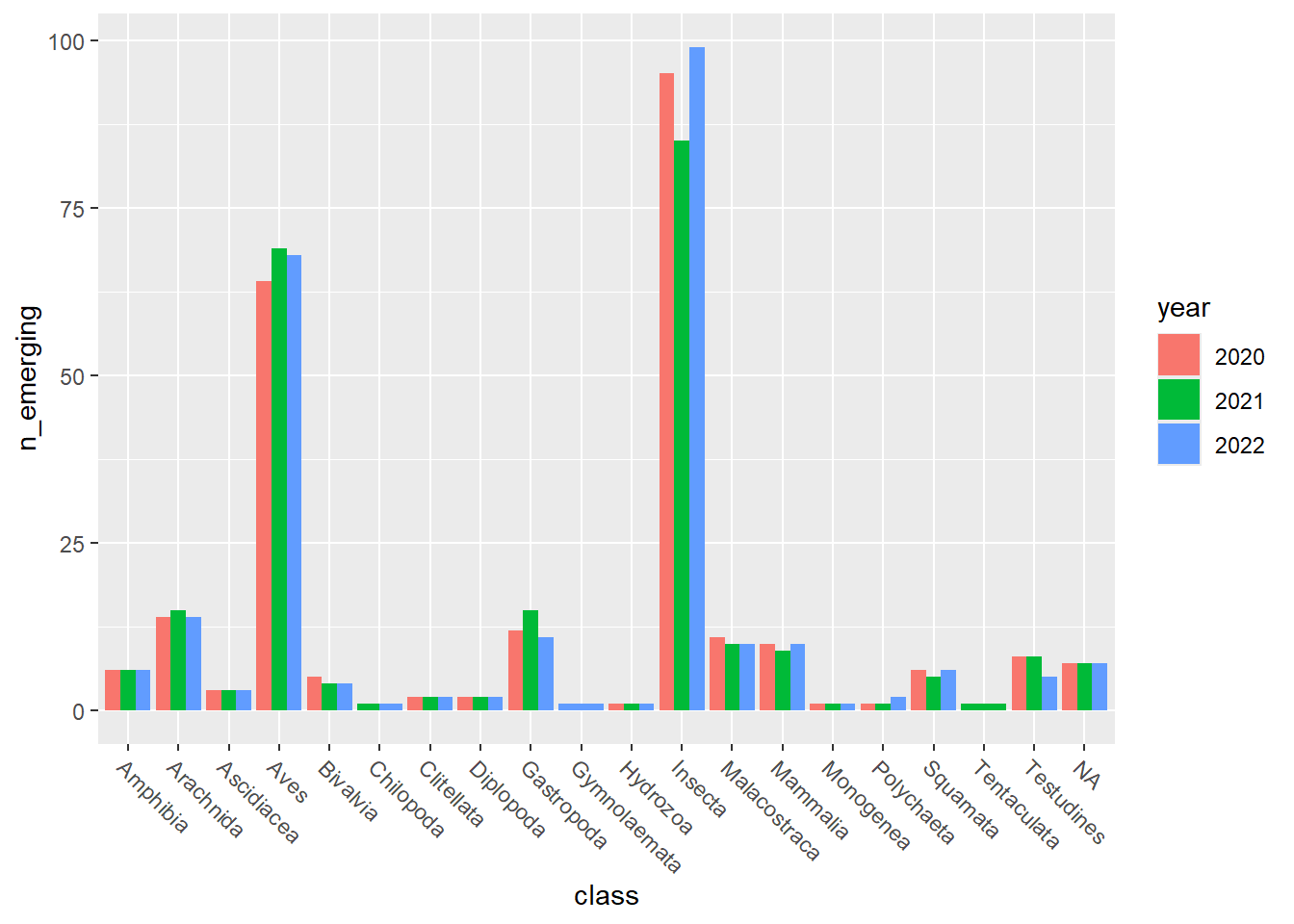
##
## [[3]]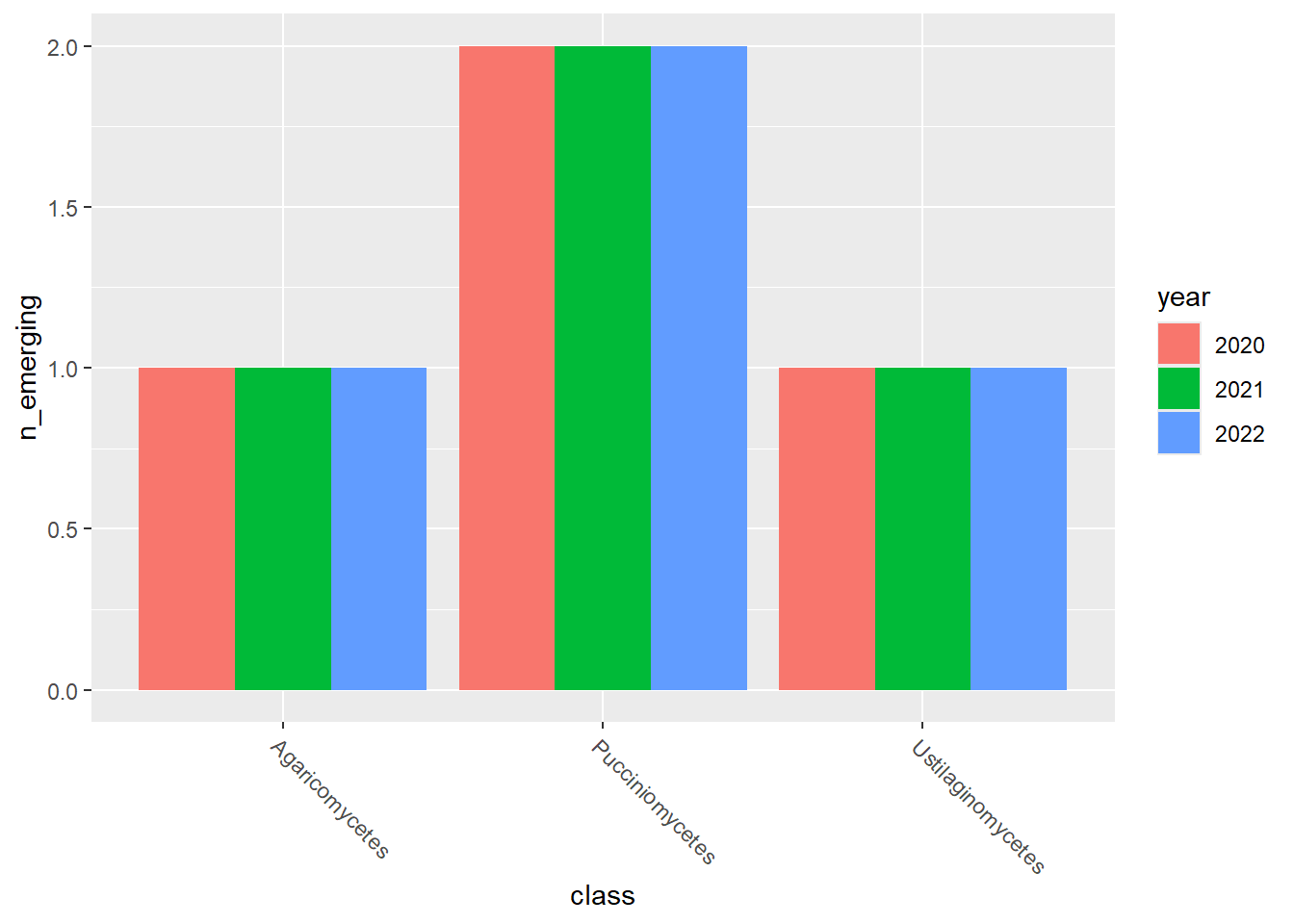
##
## [[4]]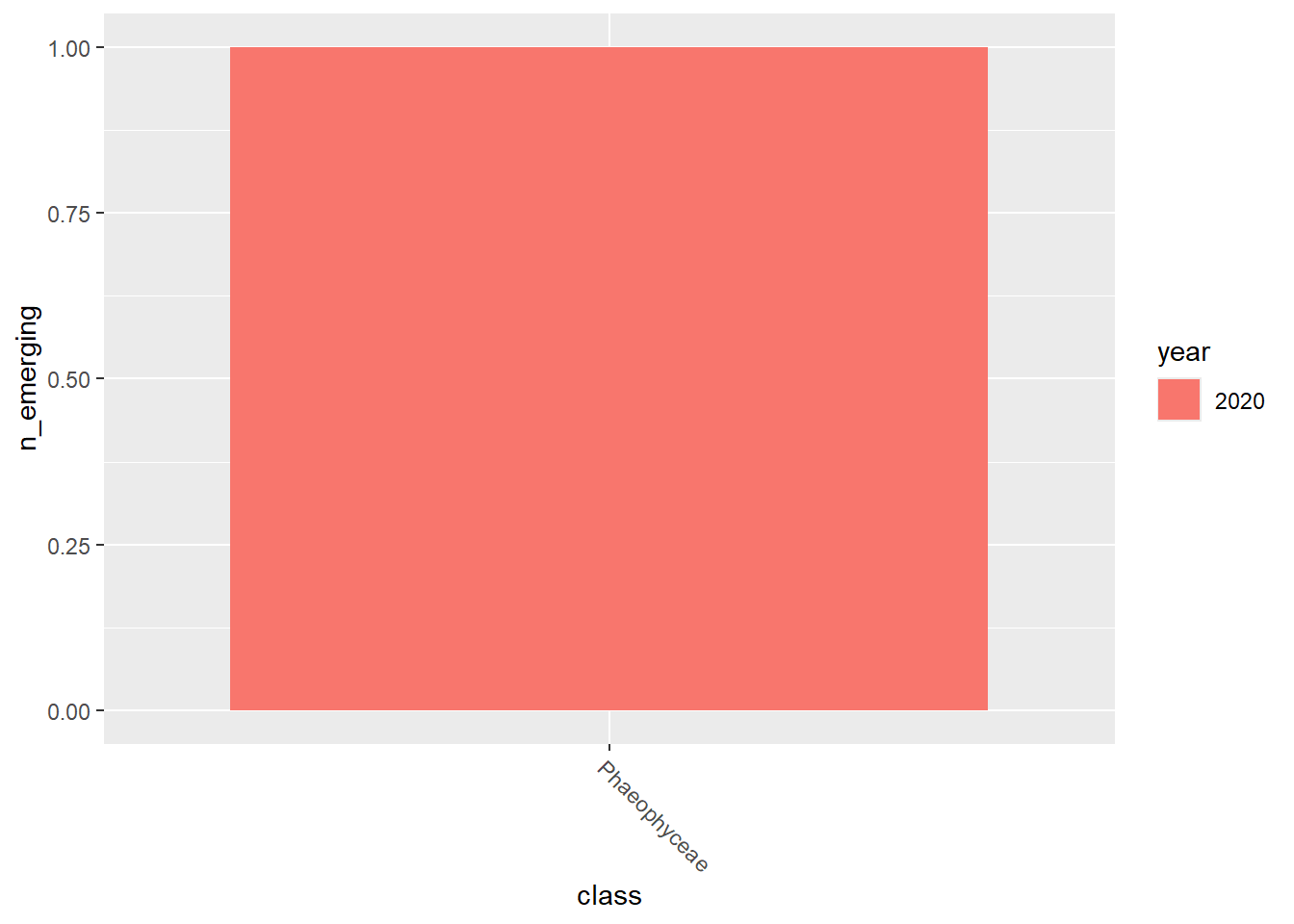
Save both table and graphs:
readr::write_tsv(
emerging_df_Natura2000_occs,
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
"n_emerging_taxa_per_class_in_Natura2000_occs.tsv"
),
na = ""
)
purrr::walk2(
emerging_plots_Natura2000_occs,
kingdoms$kingdom,
function(x, y) {
ggplot2::ggsave(
here::here(
"data",
"output",
"taxonomic_distribution_emerging_species",
paste0(
"n_emerging_taxa_per_class_in_Natura2000_occs_",
y, ".png"
)
),
x
)
}
)